
Yamini Dalal, Ph.D.
- Center for Cancer Research
- National Cancer Institute
- Building 41, Room D804B (Office)
- Bethesda, MD 20892-5055
- 240-760-6591
- dalaly@mail.nih.gov
RESEARCH SUMMARY
Our lab focuses on proteins called histones, which package the entirety of the human genome into a nucleoprotein complex called chromatin. Specific regions of chromatin are prone to fragility and chromosomal rearrangements in cancer cells. Consequently, understanding how this protein complex is mis-regulated is important in understanding chromosome fragility. Using a combination of chromatin biochemistry, computational modeling, single molecule microscopy, genetics, genomics and cell biology, we are currently investigating whether histone complexes can adopt alternate structural conformations in cancer cells, and the functional consequences of such alterations upon the cancer epigenome.
Areas of Expertise

Yamini Dalal, Ph.D.
Research
The human genome project and other sequencing projects have provided biologists with a vast amount of information about DNA sequences that comprise various organisms. In eukaryotes, the genome is intimately coupled with highly conserved canonical histone proteins, which make up half the nuclear content. Histones H2a, H2b, H3, H4 form pseudo-symmetrical bead-like units called nucleosomes around which 147bp DNA is wrapped. These repeating units package the entirety of the genome, with spacing between individual nucleosomes dictated by DNA sequence, linker histone, and non-histone proteins. Furthermore, in regions of functional importance, variant histones such as H3.3, CenH3, H2A.X, macro H2a, H2A.Z substitute their canonical counterparts within the nucleosome. These histone variants dramatically alter the structure and dynamics of the local chromatin fiber resulting in epigenetic control of functions ranging from promoter accessibility to centromere identity.
We focus on centromeres to elucidate the relationship between histone variants, chromatin fiber structure, assembly/disassembly dynamics and biological function. Centromeres are the visually striking central constriction seen in eukaryotic mitotic chromosomes, and are required for microtubule attachment. The centromeric chromatin fiber presents a near-perfect model for studying epigenetic control, because no single DNA sequence is deemed necessary for formation, specification, maintenance or function of the centromere. Rather, the key element appears to be the histone H3 variant CenH3. CenH3's exclusive enrichment at a location within the genome marks that region to be the centromere in thousands upon thousands of cell generations, providing epigenetic inheritance of centromere domains. How this remarkable feat is accomplished remains a central question in biology. We have uncovered features of native Drosophila and human CenH3-nucleosomes that differ from those seen for canonical nucleosomes in vivo. We have recently proposed that these structural differences determine the accessibility of the centromeric chromatin fiber during the cell cycle so that kinetochore proteins can distinguish CenH3 nucleosomes, and that other forms of the CenH3 nucleosome at ectopic locations that block kinetochore recognition, protecting the cell from neocentromere formation.
Current goals of the lab are to elucidate: 1) how CenH3 nucleosome structure varies through the cell cycle, 2) how assembly and disassembly kinetics influence the centromeric chromatin fiber accessibility and uniformity, 3) how the global chromatin fiber's three-dimensional arrangement and structure is altered when the individual nucleosomes that make up the fiber are replaced by CenH3 or other histone variants in colorectal tumors, 4) functional and epigenetic consequences of histone variant invasion of ectopic loci, and 5) whether the distinctive features we observe in fly and human are conserved in other species. Our collaborative projects range from developing new atomic force microscopy applications to enable fluorescent single molecule recognition within native chromatin, analyzing the structure of human artificial chromosomes, to mapping chromatin domains at fragile regions in human chromosomes.
Publications
Biography

Yamini Dalal, Ph.D.
Dr. Yamini Dalal became interested in chromosome structure and epigenetic gene regulation during her Baccalaureate years at St. Xavier's College, Bombay, India, where she graduated with a double major in Biochemistry and Life Sciences in 1995. She moved to the United States for her post-graduate work. In Arnold Stein's laboratory at Purdue University, she used classical chromatin biochemistry tools to understand how DNA sequence motifs and linker histones can shape the chromatin structure in silico, in vitro, and in vivo. During this time, she discovered that the regions of the mouse genome contained alternating tracts of stiff and flexible DNA, which allowed in silico prediction of nucleosome positions. These positions could be recapitulated in vitro using just purified histones and DNA, and detected in vivo, at developmentally regulated genes in mice. She also studied how linker histone H1 could influence nucleosome positioning and chromatin folding in vitro and in vivo. For these studies, she received her Ph.D. from Purdue University in 2003. Histone variants were the next logical step in teasing out how intrinsic variability in the chromatin fiber can encode a diversity of biological functions. To study this aspect of chromatin structure, Yamini moved to Seattle to work with Dr. Steven Henikoff at the Fred Hutchinson Cancer Research Center from 2003-2007. Her lab focuses on proteins called histones and their variants, which package the entirety of the human genome into a nucleoprotein complex called chromatin in vivo. Specific regions of chromatin are prone to fragility and chromosomal rearrangements in cancer cells. Using a combinatorial interdisciplinary toolset in colorectal and brain cancer cells and tissues, they investigate the structural and functional consequences of epigenetic alterations within the cancer epigenome, and the therapeutic potential of small molecule suppressors to target these aberrant pathways. She was awarded tenure at NIH in 2018. In 2021, Dr. Dalal became Senior Advisor for Faculty Development, CCR Office of Scientific Programs.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Team
News
News from Our Lab
Dr. Annie Gilbert and Dr. Jayeeta Kolay are 2025 FARE award recipients.
Read Dr. Bui and Dr. Dalal’s immigrant journeys in science with the American Society for Biochemistry and Molecular Biology
Read Dr. Melters and Dr. Dalal's thoughts on latest breakthroughs from HHMI Dr. Luger’s lab
Disruption of the MSL complex inhibits tumour maintenance by exacerbating chromosomal instability
Nature Cell Biology 23: 401-412, 2021
Rewiring of cellular programmes in malignant cells generates cancer-specific vulnerabilities. Here, using an unbiased screening strategy aimed at identifying non-essential genes required by tumour cells to sustain unlimited proliferative capacity, we identify the male-specific lethal (MSL) acetyltransferase complex as a vulnerability of genetically unstable cancers. We find that disruption of the MSL complex and consequent loss of the associated H4K16ac mark do not substantially alter transcriptional programmes but compromise chromosome integrity and promote chromosomal instability (CIN) that progressively exhausts the proliferative potential of cancer cells through a p53-independent This effect is dependent on pre-existing genomic instability, and normal cells are insensitive to MSL disruption. Using cell- and patient-derived xenografts from multiple cancer types, we show that excessive CIN induced by MSL disruption inhibits tumour maintenance. Our findings suggest that targeting MSL may be a valuable means to increase CIN beyond the level tolerated by cancer cells without inducing severe adverse effects in normal tissues. View Full-Text Article.
Chromatin: Nucleosomal dynamics at centromeres
Nature Reviews Genetics 13: 597, 2012
This review article by Darren J. Burgess highlights Minh Bui's research paper, "Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo" (Cell 150: 317–326, 2012) as one of two important studies that shed light on the cell-cycle-regulated stoichiometry of centromeric nucleosomes. View Full-Text Article.
Covers

High-resolution analysis of human centromeric chromatin
About the Cover:
The essential kinetochore component CENP-C is critical in regulating centromere homeostasis. The image shows a multipolar spindle metaphase cell, a consequence when CENP-C is overexpressed. Mitotic spindles (cyan) attach to the chromosomes (grey) via the kinetochore (CENP-C in red).
Abstract:
Centromeres are marked by the centromere-specific histone H3 variant CENP-A/CENH3. Throughout the cell cycle, the constitutive centromere-associated network is bound to CENP-A chromatin, but how this protein network modifies CENP-A nucleosome conformations in vivo is unknown. Here, we purify endogenous centromeric chromatin associated with the CENP-C complex across the cell cycle and analyze the structures by single-molecule imaging and biochemical assays. CENP-C complex–bound chromatin was refractory to MNase digestion. The CENP-C complex increased in height throughout the cell cycle culminating in mitosis, and the smaller CENP-C complex corresponds to the dimensions of in vitro reconstituted constitutive centromere-associated network. In addition, we found two distinct CENP-A nucleosomal configurations; the taller variant was associated with the CENP-C complex. Finally, CENP-A mutants partially corrected CENP-C overexpression–induced centromeric transcription and mitotic defects. In all, our data support a working model in which CENP-C is critical in regulating centromere homeostasis by supporting a unique higher order structure of centromeric chromatin and altering the accessibility of the centromeric chromatin fiber for transcriptional machinery.

Oncogenic lncRNAs alter epigenetic memory at a fragile chromosomal site in human cancer cells
About the Featured Article
In cancer cells, histone H3 variant CENP-A is innately overexpressed and mislocalized to regions outside centromeres, typically to chromosome arms and telomeres. Transcription and transcripts of specific oncogenic lncRNAs could serve as a recruitment signal for CENP-A mislocalization. Thus, the oncogenic lncRNA-based mechanism of CENP-A mislocalization can generate aberrant epigenetic signatures resulting in chromosome breaks and amplification in the cancer epigenome.
https://www.science.org/doi/10.1126/sciadv.abl5621
Abstract
Chromosome instability is a critical event in cancer progression. Histone H3 variant CENP-A plays a fundamental role in defining centromere identity, structure, and function but is innately overexpressed in several types of solid cancers. In the cancer background, excess CENP-A is deposited ectopically on chromosome arms, including 8q24/cMYC locus, by invading transcription-coupled H3.3 chaperone pathways. Up-regulation of lncRNAs in many cancers correlates with poor prognosis and recurrence in patients. We report that transcription of 8q24-derived oncogenic lncRNAs plays an unanticipated role in altering the 8q24 chromatin landscape by H3.3 chaperone–mediated deposition of CENP-A–associated complexes. Furthermore, a transgene cassette carrying specific 8q24-derived lncRNA integrated into a naïve chromosome locus recruits CENP-A to the new location in a cis-acting manner. These data provide a plausible mechanistic link between locus-specific oncogenic lncRNAs, aberrant local chromatin structure, and the generation of new epigenetic memory at a fragile site in human cancer cells.
SCIENCE ADVANCES, 2 Mar 2022, Vol 8, Issue 9. DOI: 10.1126/sciadv.abl5621
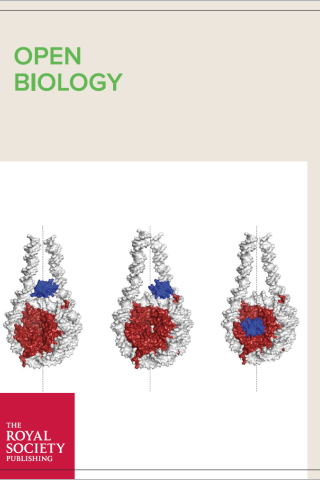
A Glitch in the Snitch: the role of linker histone H1 in shaping the epigenome in normal and diseased cells
About the cover
Schematic representation of H1 binding modes. Image credit: Ankita Saha and Yamini Dalal.
Taken from Open Biology article “A Glitch in the Snitch: the role of linker histone H1 in shaping the epigenome in normal and diseased cells” DOI: 10.1098/rsob.210124
Abstract
Histone H1s or the linker histones are a family of dynamic chromatin compacting proteins that are essential for higher-order chromatin organization. These highly positively charged proteins were previously thought to function solely as repressors of transcription. However, over the last decade, there is a growing interest in understanding this multi-protein family, finding that not all variants act as repressors. Indeed, the H1 family members appear to have distinct affinities for chromatin and may potentially affect distinct functions. This would suggest a more nuanced contribution of H1 to chromatin organization. The advent of new technologies to probe H1 dynamics in vivo, combined with powerful computational biology, and in vitro imaging tools have greatly enhanced our knowledge of the mechanisms by which H1 interacts with chromatin. This family of proteins can be metaphorically compared to the Golden Snitch from the Harry Potter series, buzzing on and off several regions of the chromatin, in combat with competing transcription factors and chromatin remodellers, thereby critical to the epigenetic endgame on short and long temporal scales in the life of the nucleus. Here, we summarize recent efforts spanning structural, computational, genomic and genetic experiments which examine the linker histone as an unseen architect of chromatin fibre in normal and diseased cells and explore unanswered fundamental questions in the field.
Saha Ankita and Dalal Yamini 2021 A glitch in the snitch: the role of linker histone H1 in shaping the epigenome in normal and diseased cells Open Biol. 11210124210124 http://doi.org/10.1098/rsob.210124

Diving into Chromatin Across Time and Space
About the Cover:
This special issue “Diving into chromatin structure across space and time” highlights the application of cutting-edge approaches to further illuminate transactions that occur on, and by, the chromatin fiber. Appropriately for this year of covid, where virtual life is a simulacrum of reality, we start at one end of the scale - computational modeling of dynamic nucleoprotein complexes.
Dalal Y, Panchenko AR. Diving into Chromatin across Space and Time. J Mol Biol. 2021 Mar 19;433(6):166884. doi: 10.1016/j.jmb.2021.166884. Epub 2021 Feb 20. PMID: 33621519.

Promiscuous histone mis-assembly is actively prevented by chaperones
About the Cover
Chaperone HJURP drives the proper loading of protein CENP-A to the centromere of a chromosome. The effect of HJURP on CENP-A's structural dynamics are observed and explained using dual-resolution in silico simulations, while in vivo experiments demonstrate how CENP-A mutations influence its specific localization in human cells.
Abstract
Histone proteins are essential for the organization, expression, and inheritance of genetic material for eukaryotic cells. A centromere-specific H3 histone variant, centromere protein A (CENP-A), shares about 50% amino acid sequence identity with H3. CENP-A is required for packaging the centromere and for the proper separation of chromosomes during mitosis. Despite their distinct biological functions, previously reported crystal structures of the CENP-A/H4 and H3/H4 dimers reveal a high degree of similarity. In this work, we characterize the structural dynamics of CENP-A/H4 and H3/H4 dimers based on a dual-resolution approach, using both microsecond-scale explicit-solvent all-atom and coarse-grained (CG) molecular dynamics (MD) simulations. Our data show that the H4 histone is significantly more rigid compared with the H3 histone and its variant CENP-A, hence, serving as a reinforcing structural element within the histone core. We report that the CENP-A/H4 dimer is significantly more dynamic than its canonical counterpart H3/H4, and our results provide a physical explanation for this flexibility. Further, we observe that the centromere-specific chaperone Holliday Junction Recognition Protein (HJURP) stabilizes the CENP-A/H4 dimer by forming a specific electrostatic interaction network. Finally, replacing CENP-A S68 with E68 disrupts the binding interface between CENP-A and HJURP in all-atom MD simulation, and consistently, in vivo experiments demonstrate that replacing CENP-A S68 with E68 disrupts CENP-A’s localization to the centromere. Based on all our results, we propose that, during the CENP-A/H4 deposition process, the chaperone HJURP protects various substructures of the dimer, serving both as a folding and binding chaperone.
Promiscuous histone mis-assembly is actively prevented by chaperones. Zhao, Haiqing+; Winogradoff, David+; Bui, Minh; Dalal, Yamini*; and Papoian, Garegin*. J Am Chem Soc. 138(40): 13083-13446, 2016. +equal contribution; *corresponding authors. Note: Haiqing Zhao is a joint graduate student who was funded for 2 years by the NCI-University of Maryland Partnership for Integrative Cancer Research.







