Tumor Virus RNA Biology Section
Zhi-Ming Zheng, M.D., Ph.D.
Research
Dr. Zheng’s lab, the Tumor Virus RNA Biology Section, has been studying protein-RNA interactions and their consequences in various infections with tumor viruses, including high-risk human papillomaviruses and Kaposi’s sarcoma-associated herpesvirus. The aim is to understand how RNA splicing and regulatory RNAs regulate the expression of viral and host genes in viral carcinogenesis. The long-term goal is to develop a series of therapeutic approaches to control viral or cellular gene expression for cancer or AIDS treatments and to identify biomarkers for clinical diagnosis and prognosis.
Team
Covers
Induction of translation-suppressive G3BP1+ stress granules and interferon-signaling cGAS condensates by transfected plasmid DNA
Plasmid DNA transfection is one of the fundamental tools of biomedical research. Here, we found that plasmid DNA transfection mediated by liposomes activates multiple innate immune responses in several widely used cell lines. Their activations were visible by the detection of stress granules (SG) and cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS)-DNA condensates (cGC) in the transfected cells in a plasmid DNA dose-dependent manner. The elevated levels of phosphorylated eukaryotic translation initiation factor 2 subunit alpha (eIF2α), interferon regulatory factor 3 (IRF3), and signal transducer and activator of transcription 1 (STAT1) were induced in plasmid DNA-transfected cells. The formation of SG but not cGC required active transcription and the formation of double-stranded RNA in transfected cells. Plasmid DNA-induced SG or cGC were mutually exclusive because they triggered two distinct pathways. Knockdown (KD) of protein kinase R (PKR) before plasmid DNA transfection led to abolishing SG without affecting cGC formation. Conversely, cGAS KD could prevent cGC without affecting SG formation. In addition, plasmid DNA-induced SG and cGC formation could be prevented, respectively, by co-expression of Kaposi's sarcoma-associated herpesvirus proteins ORF57 (PKR inhibitor) and ORF52 (cGAS inhibitor). Inhibition of SG formation mediated by PKR KD, but not cGC KD, also led to increased expression of transgenes, indicating that PKR activation represents a major roadblock to gene expression. Together, these data indicate that plasmid DNA triggers innate immune responses in the transfected cells and causes a significant cellular perturbation that should be considered during experiment design and data interpretation.
Majerciak V, Zheng ZM. Induction of translation-suppressive G3BP1+ stress granules and interferon-signaling cGAS condensates by transfected plasmid DNA. hLife 2025;3:21–37

Revisiting and corrections to the annotated SRSF3 (SRp20) gene structure and RefSeq sequences from the human and mouse genomes
SRSF3 (SRp20) is the smallest member of the serine/arginine (SR)-rich protein family. We found the annotated human SRSF3 and mouse Srsf3 RefSeq sequences are much larger than the detected SRSF3/Srsf3 RNA size by Northern blot. Mapping of RNA-seq reads from various human and mouse cell lines to the annotated SRSF3/Srsf3 gene illustrated only a partial coverage of its terminal exon 7. By 5ʹ RACE and 3ʹ RACE, we determined that SRSF3 gene spanning over 8422 bases and Srsf3 gene spanning over 9423 bases. SRSF3/Srsf3 gene has seven exons with exon 7 bearing two alternative polyadenylation signals (PAS). Through alternative PAS selection and exon 4 exclusion/inclusion by alternative RNA splicing, SRSF3/Srsf3 gene expresses four RNA isoforms. The major SRSF3 mRNA isoform with exon 4 exclusion by using a favorable distal PAS to encode a full-length protein is 1411 nt long (not annotated 4228 nt) and the same major mouse Srsf3 mRNA isoform is only 1295 nt (not annotated 2585 nt). The difference from the redefined RNA size of SRSF3/Srsf3 to the corresponding RefSeq sequence is at the 3’ UTR region. Collectively, the redefined SRSF3/Srsf3 gene structure and expression will allow better understanding of SRSF3 functions and its regulations in health and diseases.
Lulu Yu, Vladimir Majerciak, Rong Jia, Zhi-MIng Zheng, Revisiting and corrections to the annotated SRSF3 (SRp20) gene structure and RefSeq sequences from the human and mouse genomes. Cell Insight 2 (2): e100089, 2023. https://doi.org/10.1016/j.cellin.2023.100089

Towards Better Understanding of KSHV Life Cycle: from Transcription and Posttranscriptional Regulations to Pathogenesis 2) HPV18 Utilizes Two Alternative Branch Sites for E6*I Splicing to Produce E7 Protein
1) Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus-8 (HHV-8), is etiologically linked to the development of Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. In this review, Yan and Majerciak et al. summarize the current research status on the biology of latent and lytic viral infection, the regulation of viral life cycles and the related pathogenesis.
2) Human papillomavirus 18 (HPV18) E6 and E7 oncogenes are transcribed as a single bicistronic E6E7 pre-mRNA. The E6 ORF region in the bicistronic E6E7 pre-mRNA contains an intron. Splicing of this intron disrupts the E6 ORF integrity and produces a spliced E6*I RNA for efficient E7 translation. Brant et al. identified two alternative branch sites in the HPV18 E6 intron which selection correlates to the efficiency of E6*I splicing and the production of E6 and E7 oncoproteins during productive or oncogenic HPV infection.
1) Lijun Yan, Vladimir Majerciak, Zhi-Ming Zheng, Ke Lan. Towards Better Understanding of KSHV Life Cycle: from Transcription and Posttranscriptional Regulations to Pathogenesis. Virologica Sinica 34 (2):135–161, 2019. https://doi.org/10.1007/s12250-019-00114-3
2) Ayslan Castro Brant, Vladimir Majerciak, Miguel Angelo Martins Moreira, Zhi-Ming Zheng. HPV18 Utilizes Two Alternative Branch Sites for E6*I Splicing to Produce E7 Protein. Virologica Sinica 34 (2): 211–221, 2019. https://doi.org/10.1007/s12250-019-00098-0
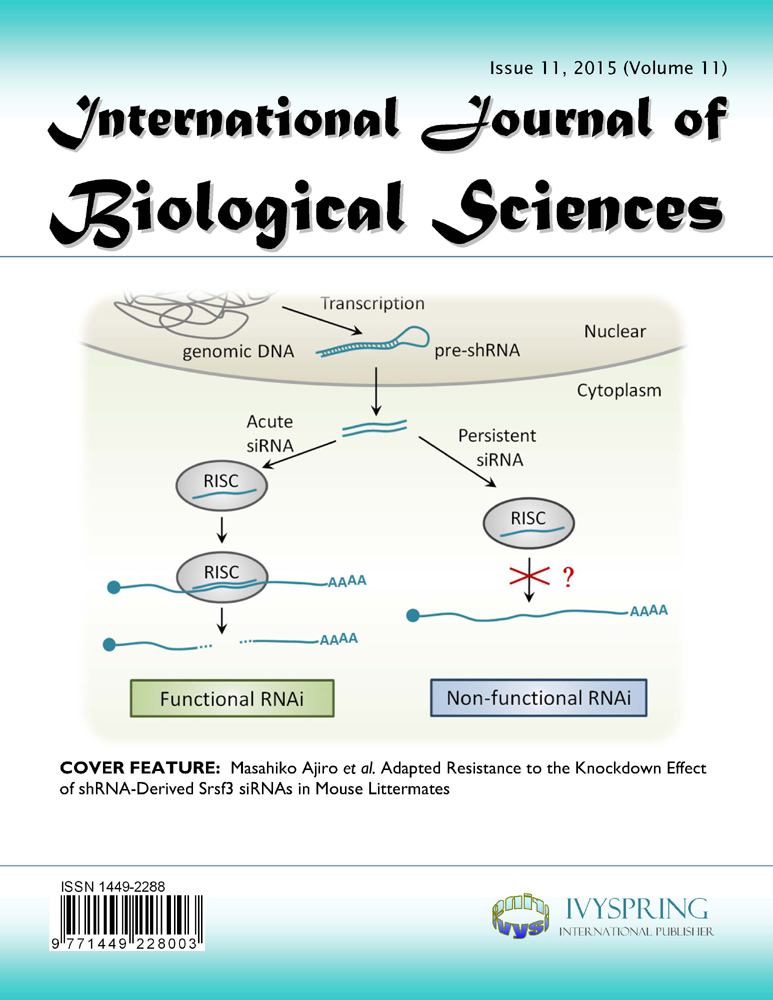
Adapted Resistance to the Knockdown Effect of shRNA-Derived Srsf3 siRNAs in Mouse Littermates
Gene silencing techniques are widely used to control gene expression and have potential for RNAi-based therapeutics. In this report, transgenic mouse lines were created for conditional knockdown of Srsf3 (SRp20) expression in liver and mammary gland tissues by expressing Srsf3-specific shRNAs driven by a U6 promoter. Although a small portion of the transgenic mouse littermates were found to produce siRNAs in the targeted tissues, most of the transgenic littermates at two months of age failed to display a knockdown phenotype of Srsf3 expression in their liver and mammary gland tissues where an abundant level of Srsf3 siRNAs remained. We saw only one of four mice with liver/mammary gland expressing Srsf3 siRNA displayed a suppressed level of Srsf3 protein, but not the mRNA. Data indicate that the host resistance to a gene-specific siRNA targeting an essential gene transcript can be developed in animals, presumably as a physiological necessity to cope with the hostile perturbation.
Ajiro M, Jia R, Wang RH, Deng CX, Zheng ZM. Adapted Resistance to the Knockdown Effect of shRNA-Derived Srsf3 siRNAs in Mouse Littermates. Int J Biol Sci 11(11):1248-1256, 2015. doi:10.7150/ijbs.13011

Kaposi's Sarcoma-Associated Herpesvirus
The discovery of KSHV in 1994 was a historical landmark in tumor virology and human cancer research. KSHV's subsequent identification as a cause of Kaposi sarcoma and its association with primary effusion lymphoma and multicentric Castleman disease soon attracted the attention of hundreds of research laboratories and motivated thousands of virologists and oncologists to switch their research directions. To date, PubMed has collected nearly 5000 papers on KSHV from numerous journal publications throughout the world. These studies indicate that the global fight against human cancers will continue to receive great support from our tremendous efforts in searching for new tumor-causing viruses and in understanding the basic biology of tumor viruses. To celebrate the 20th year of KSHV's discovery, I am very proud to be an invited Guest Editor for a Special Issue on KSHV in Viruses and am happy to assemble all published articles from the Special Issue into this book, Kaposi Sarcoma Associated Herpesvirus.
This book is a reprint of the special issue that appeared in the online open access journal Viruses (ISSN 1999-4915) in 2014.
Job Vacancies
We have no open positions in our group at this time, please check back later.
To see all available positions at CCR, take a look at our Careers page. You can also subscribe to receive CCR's latest job and training opportunities in your inbox.
Alumni
Lab Life

Ayslan Castro Brant, Ph.D., Postdoctoral Fellow (Visiting)
ayslan.castrobrant@nih.gov
Ayslan earned his Ph.D. in genetics from Rio de Janeiro Federal University (UFRJ), Brazil, under the mentorship of Dr. Miguel Moreira, Brazilian National Cancer Institute (INCA) and co-mentorship of Dr. Zhi-Ming Zheng (HIV DRP). His work is focused on understanding how SARS-CoV-2 synthesizes its subgenomic RNAs (sgRNAs) for virus replication. Recently Ayslan received a travel award from and was invited to serve as a session co-chair at the International Papillomavirus Conference (IPVC2020) in Barcelona.
Deanna Gotte, M.S., Research Biologist
gotted@mail.nih.gov
Deanna earned her M.S. in applied microbiology from Northern Arizona University. She provides laboratory support for the Zheng lab. Her current research is focused on understanding the role of nonsense-mediated RNA decay in human papillomavirus (HPV) RNA processing and half-life. Deanna’s research career has spanned over many disciplines including retroviral proteases, HIV-1 subtype analysis, and transcription fidelity of Saccharomyces cerevisiae RNA polymerase II.

Vladimir Majerciak, Ph.D., Associate Scientist
majerciv@mail.nih.gov
Vladimir earned his Ph.D. in virology from Comenius University and the Institute of Virology, Slovak Academy of Sciences in Bratislava, Slovakia. His current research is mainly focused on Kaposi’s sarcoma-associated herpesvirus (KSHV) and KSHV RNA-binding protein ORF57. Recently, he has expanded his study to include other tumor viruses, with a focus on virus-host interactions, viral protein structure, and viral transcriptome profiling. He was promoted to Staff Scientist in 2011 and to Associate Scientist in 2019. Read more about Vladimir in his CCR scientist profile.
Lulu Yu, Ph.D., Research Fellow
lulu.yu@nih.gov
Lulu earned her Ph.D. in cancer epidemiology from Peking Union Medical College, China, where she conducted human papillomavirus (HPV) epidemiology studies on cervical cancer in the general population under the mentorship of Dr. Youlin Qiao. Currently she is studying how papillomaviruses and the cellular splicing factor SRSF3 (SRp20) regulate RNA-binding protein expression, RNA splicing and other RNA processing, and oncogenesis. She is a winner of the NIH Fellows Award for Research Excellence (FARE) in 2019 and the ASV Travel Award in 2020.

2024
Left to right: Zhi-Ming Zheng, Lulu Yu, Deanna Gotte, Angelika Chen, Vladimir Majerciak, Ayslan Castro Brant, Md Nazmul Hossain, Jian Yin

2023
Front row (left-right): Lulu Yu, Deanna Gotte, Zhi-Ming (Thomas) Zheng
Back row (left-right): Shivalee Duduster, Ayslan Castro Brant, Vladimir Majerciak, Sukkum Chang
2020
Top row (left to right): Zhi-Ming Zheng and his wife, Vladimir Majerciak, Haibin Liu and his wife and son
Bottom row (left to right): Deanna Gotte and her husband, Lulu Yu and her husband, Beatriz Alvarado Hernandez and her husband
Not shown: Ayslan Brant and his wife

2019
Left to right: Haibin Liu, Deanna Gotte, Beatriz Hernandez, Thomas Zheng, Mina Griffioen, Lulu Yu, Vladimir Majerciak

2018
Front row (left to right): Mina Griffioen, Beatriz Hernandez, Vladimir Majerciak
Back row (left to right): Thomas Zheng, Deanna Gotte, Lulu Yu, Haibin Liu

2017
Front row (left to right): Tingtine Zhang, Lulu Yu, Beatriz Hernandez, Vladimir Majerciak, Ayslan Brant
Back row (left to right): Deanna Gotte, Thomas Zheng, Andrew BeltCappellino, Pengfei Jiang, Haibin Liu

2016
Front row: Vladimir Majerciak
Back row (left to right): Thomas Zheng, Nishi Sharma, Haibin Liu, Masahiko Ajiro, Xiang Yang Xue

2015
Left to right: Masahiko Ajiro, Yanping Ma, Junfen Xu, Echo Zhang, Thomas Zheng, Vincent Homman, Vladimir Majerciak, Nishi Sharma

2014
Front row (left to right): Vincent Homman, Yanping Ma, Xiaohong Wang, Junfen Xu
Back row (left to right): Nishi Sharma, Vladimir Majerciak, Thomas Zheng, Masahiko Ajiro

2013
Front row (left to right): Vladimir Majerciak, Nishi Sharma, Xiaohong Wang
Back row (left to right): Masahiko Ajiro, Xiaofan Li, Thomas Zheng

2012

2011

2010

2009

2008

2007

2006

2004

2002

































