Innovative imaging methods developed and refined within CCR revealed atomic-level structures of biological molecules and unveiled dynamic views of a cell’s interior that are driving the design of new treatments and diagnostics for cancer.
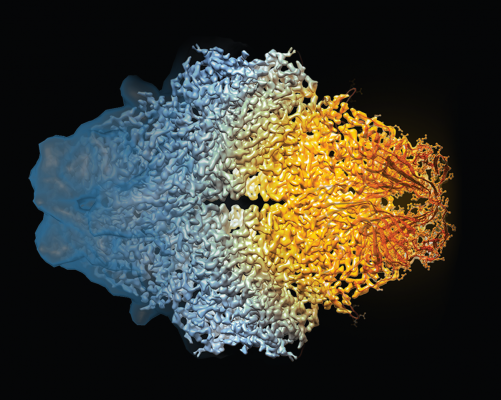
This composite image of the beta-galactosidase molecule shows how cryo-EM's resolution has dramatically improved in recent years.
Credit: Veronica Falconieri, Sriram Subramaniam, CCR, NCI, NIH
A unique aspect of CCR is an interest in the development of new methods and technology to drive discovery. Innovative imaging methods developed and refined within CCR have revealed atomic-level structures of biological molecules and unveiled dynamic views of a cell’s interior. These technologies have enabled groundbreaking discoveries at the subcellular level that are driving the design of new treatments and diagnostics for cancer.
For decades, X-ray crystallography has been the primary mainstay of structural studies of biological molecules. The technique has yielded molecular structures at a very high resolution but only worked optimally with molecules that could be crystallized without destroying the integrity of the molecule itself. CCR investigator Sriram Subramaniam, Ph.D., and his team have now made it possible to obtain the same type of high-resolution structural information for many molecules that are incompatible with X-ray crystallography through his use of cryo-electron microscopy, or cryo-EM. Cryo-EM could become an important part of accelerating drug discovery. It was named “Method of the Year” by Nature in 2015 and in 2017, was recognized by the Nobel Prize in Chemistry.
To visualize molecules with cryo-EM, purified proteins are flash-frozen in liquid nitrogen and bombarded with electrons to capture a molecular image. In 2015, the Subramaniam laboratory reported the highest-resolution image ever produced with the technique, visualizing the structure of a bacterial enzyme with a level of detail previously achieved only with X-ray crystallography. Since then, his group has produced extraordinarily high-resolution structures that offer insight into how potential targets for new cancer therapies work and interact with compounds that block their function. This type of information is expected to help guide the design of new drugs for patients.
Diagnostic labs around the world rely on a method developed at CCR to visualize chromosomes and diagnose disease. The method, developed by Thomas Ried, M.D., in 1997, is called spectral karyotyping (SKY). It paints different chromosomes with distinctly colored fluorescent probes so that each one can be easily identified. SKY is particularly useful in cancer cells, whose broken and mixed up chromosomes can make other methods of chromosome identification nearly impossible.
To fully understand a biological molecule’s involvement in health or disease, scientists also need to understand how it behaves in its natural context. In the early 2000s, the laboratory of Tom Misteli, Ph.D., started devising methods to visualize how genes behave in living cells. The group developed techniques using fluorescent tags to observe gene-regulating proteins in action and used the tags to show that the protein activities are surprisingly dynamic. They also created complementary approaches to track the locations and motion of genes themselves. Those tools led to the discovery that genes have particular three-dimensional positions within the cell nucleus and their spatial organization has a profound impact on their level of activity. In 2009, Misteli’s team discovered that some genes reside in different places inside cancer cells than in noncancerous cells. Following these displaced genes may one day help clinicians diagnose disease.
The imaging and visualization methods developed by CCR scientists have propelled studies of the mechanisms that drive cancer and have influenced imaging as well as other aspects of diagnostic and clinical areas of oncology. As researchers have proven, when the unseen becomes visible, our understanding of biology gains sharper focus.
Reference
- Schröck E, et al. Science. 1996;273(5274):494-7.
- Liyanage M, et al. Nat Genet. 1996;14(3):312-5.
- Phair and Misteli, Nature, 2000;404(6778):604-9.
- Meaburn KJ, et al. J Cell Biol. 2009;187(6):801-1.
- Roukos V, et al. Science. 2013;341(6146):660-4.
- Bartesaghi A, et al. Science. 2015;348(6239):1147-51.
- Meyerson JR, et al. Nature. 2016;537(7621):567-571.


